Back to top
De Novo Sequencing

- Assemble a genome of a genetically uncharacterized species
- Detect undiscovered genes
Overview
Considerations before starting a de novo sequencing project:
- DNA quality and putative contaminations?
- Illumina or PacBio?
- Read length?
- Library size?
- Sequencing depth?
- Taxonomic neighbourhood?
- Follow-up studies?
Let us guide you – from design to analysis
Example projects using de novo sequencing:
- De novo sequencing assembly of previous uncharacterized halophile bacteria
- De novo assembly of a known but strongly mutated fungi
- Building a draft reference genome for an agricultural important plant
- Assembly of unmapped reads within a resequencing project (new plasmids, large insertions)
- Frequent bacterial de novo sequencing
Applications related to de novo sequencing:
- Reference transcriptome generation
- Resequencing
Workflow

For further reading and a detailed technical description, please download our Application Note Illumina de novo Sequencing (see related downloads).
Results
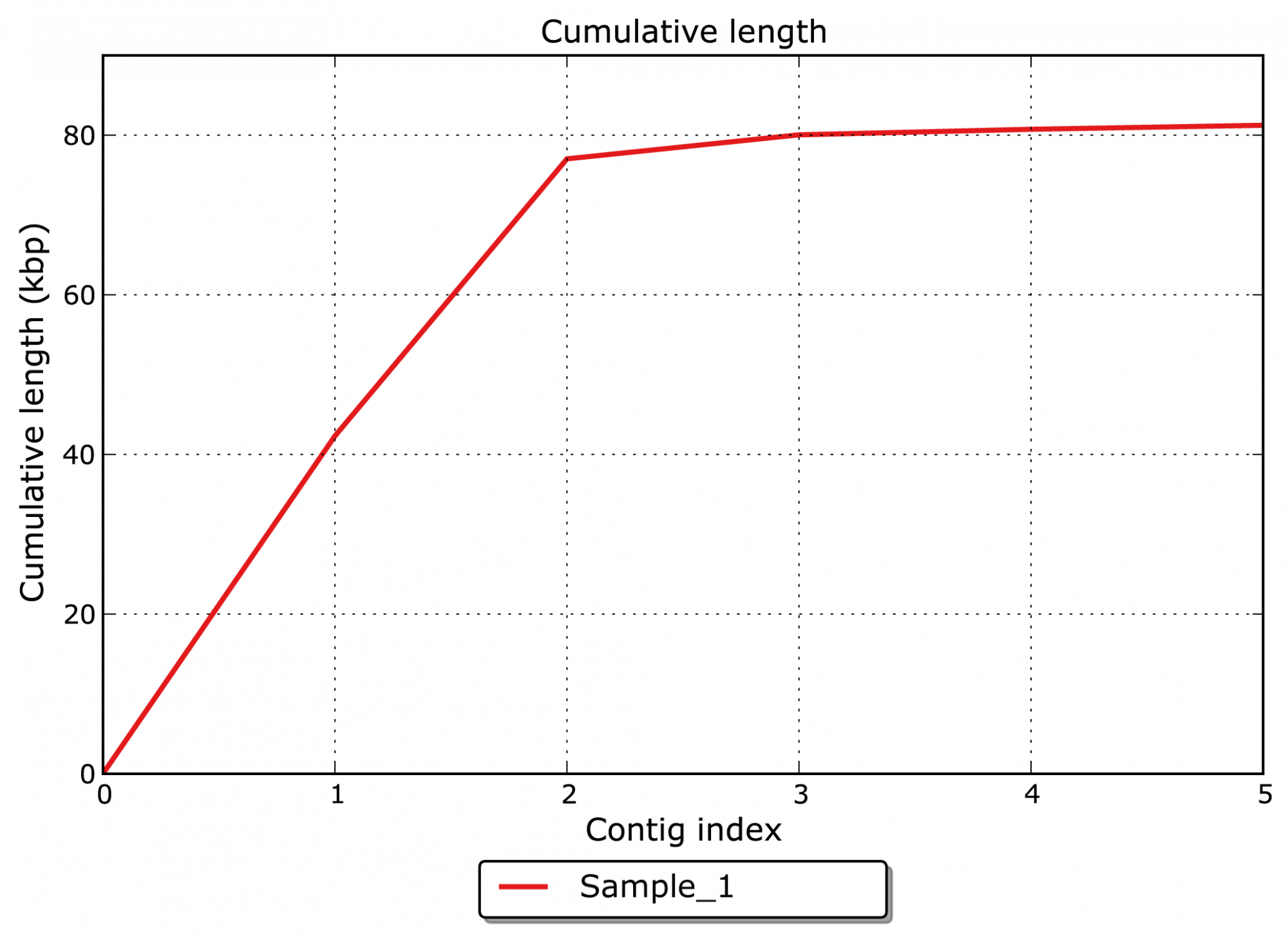
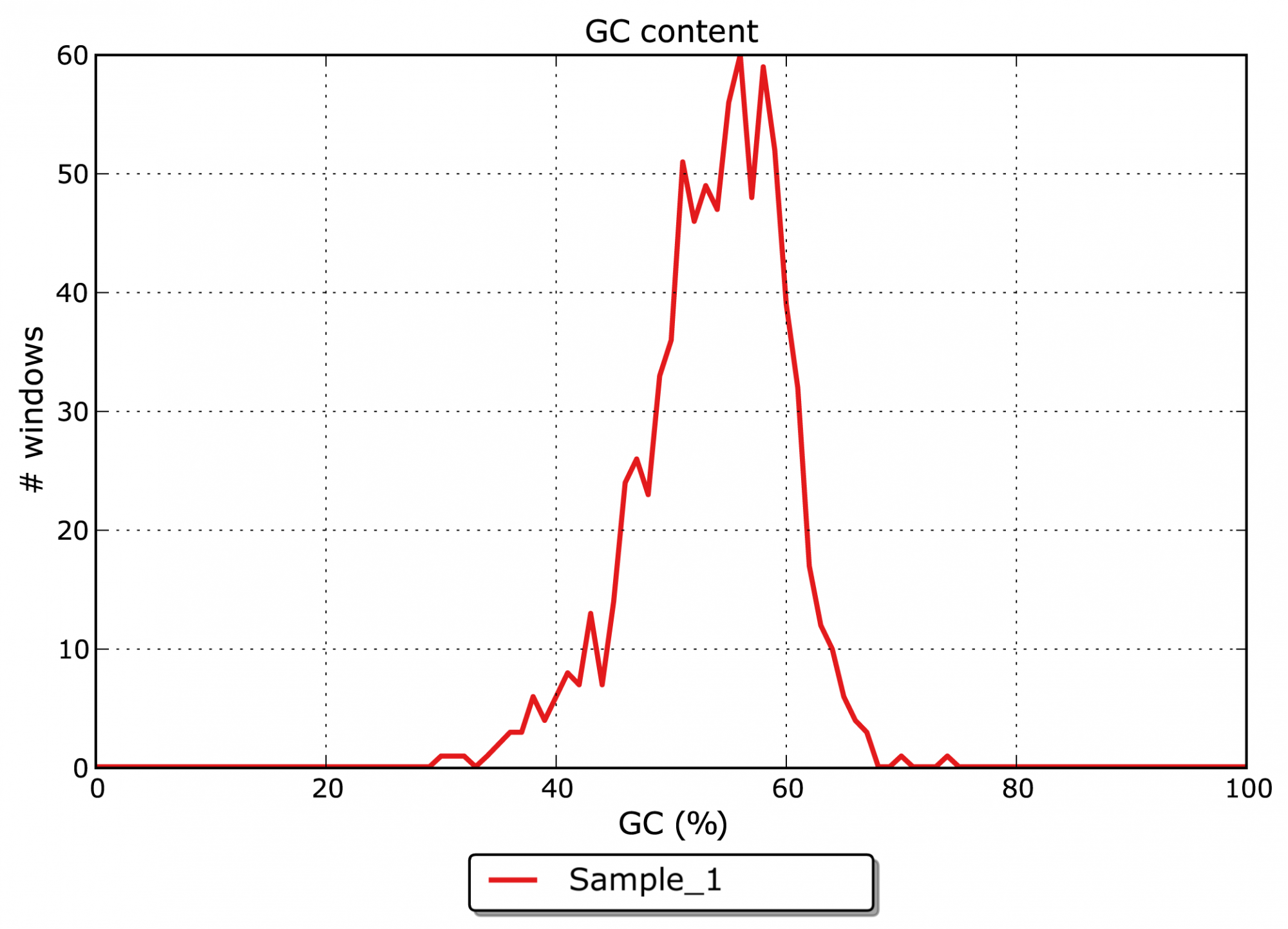
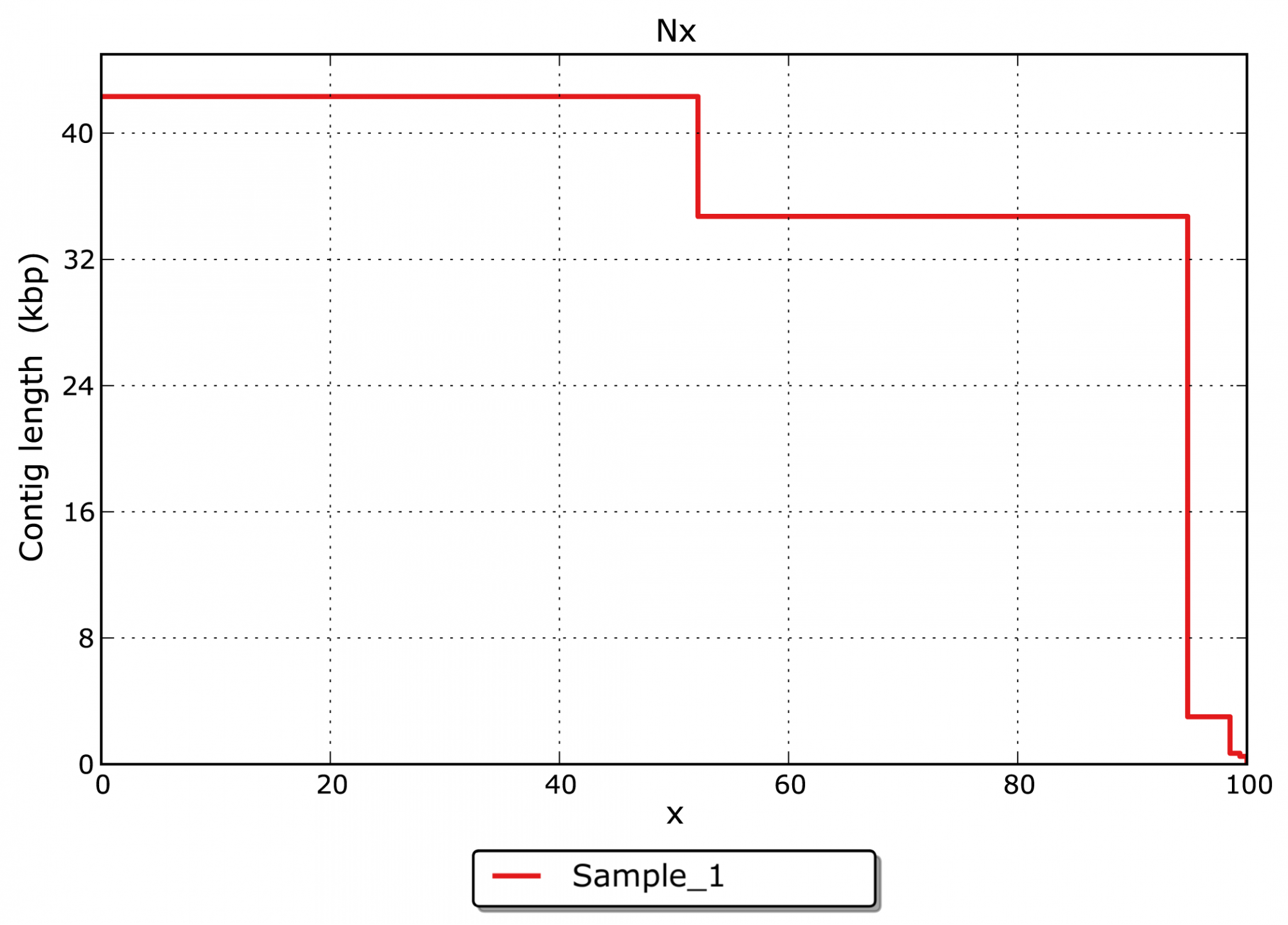
The major goal of an assembly is to merge single reads from NGS or Sanger sequencing to larger contigs and whole genome sequences. The assembly allows insights into the genomic structure of an organism and the organization of its genetic content.
Our assembly module can process reads derived from all kind of organisms including bacterial genomes as well as more complex eukaryotic organisms. Both, de novo approaches and reference guided assemblies are possible.
The module results in the assembled contigs along with statistics that answer the following questions:
- What is the overall cumulative length of the assembled contigs? (see Figure 1)
- What is the GC content of the assembled contigs? (see Figure 2)
- What is the size distribution for the assembled contigs? (see Figure 3)
Turnaround Time
- Delivery of data within 20 working days upon sample receipt (includes library preparation and sequencing)
- Additional 15 working days for data analysis (bioinformatics)
- Express service possible on request