Back to top
Séquençage de l'ARNm pour l'analyse de l'expression différentielle des gènes

- Comparer des profils d'expression génétique obtenus dans des conditions différentes
- Étudier les profils génétiques du niveau de la transcription à celui de la voie de transmission
Aperçu
Considérations avant de commencer un projet de séquençage d'ARNm :
- Objectif scientifique
- Matériel de départ et services souhaités (de l'isolement à la bioinformatique)
- Organisme et fraction d'ARN (enrichissement en poly(A), déplétion en ribo)
- Données disponibles (organisme modèle, génome de référence)
- Profondeur de séquençage (sensibilité)
- Répliques (confiance)
- Longueur de séquençage (spécificité)
- Quantités d'échantillons (complexité)
- Dispositif expérimental (nombre d'échantillons, de réplicats, de conditions à comparer)
Laissez-nous vous guider - de la conception à l'analyse
Exemples de projets utilisant le séquençage de l'ARNm :
- Études des protéines fonctionnelles et des voies moléculaires
- Des changements dans l'expression des gènes provoqués par une maladie
- Expériences liées à la perte, le gain ou au rétablissement d'une fonction
- Partie d'un projet de caractérisation en sciences omiques
- Changements fonctionnels dus aux interactions entre les espèces
- Découverte de nouveaux gènes ou d'ARN régulateurs non codants
- Détection des variants d'ARN
- Tests de médicaments
Demandes relatives au séquençage de l'ARNm :
- Shotgun métatranscriptomique
- Séquençage de petits ARN
Déroulement
Résultats
Sans Bioinformatique
Données brutes
Si aucun module d'analyse bioinformatique n'est commandé, Microsynth fournit pour le séquençage de l'ARNm les résultats clés listés ci-dessous :
- Évaluation de la quantité et de la qualité du séquençage (au format .xlsx)
Évaluer la quantité et la qualité des données de séquençage. - Données brutes (par échantillon, au format .fastq)
Accéder aux données brutes pour une analyse personnalisée ou une référence. - Rapport de synthèse du projet (format .pdf)
Résume les paramètres clés du projet.
Avec Bioinformatique
Analyse bioinformatique standard
Pour notre application de séquençage d'ARNm, le module de Microsynth fournit une multitude d'informations, répondant à vos objectifs scientifiques :
Analyse d'expression :
- Rapport complet (format .html interactif)
Approfondir les données, trier et filtrer les résultats de manière interactive. - Fichiers d'alignement/de cartographie et index (aux formats .bam et .bai)
Accédez aux fichiers d'alignement/de cartographie ainsi qu'aux index correspondants pour retracer chaque lecture jusqu'au génome de référence. - Nombre de lectures par rapport aux caractéristiques annotées (au format .tsv)
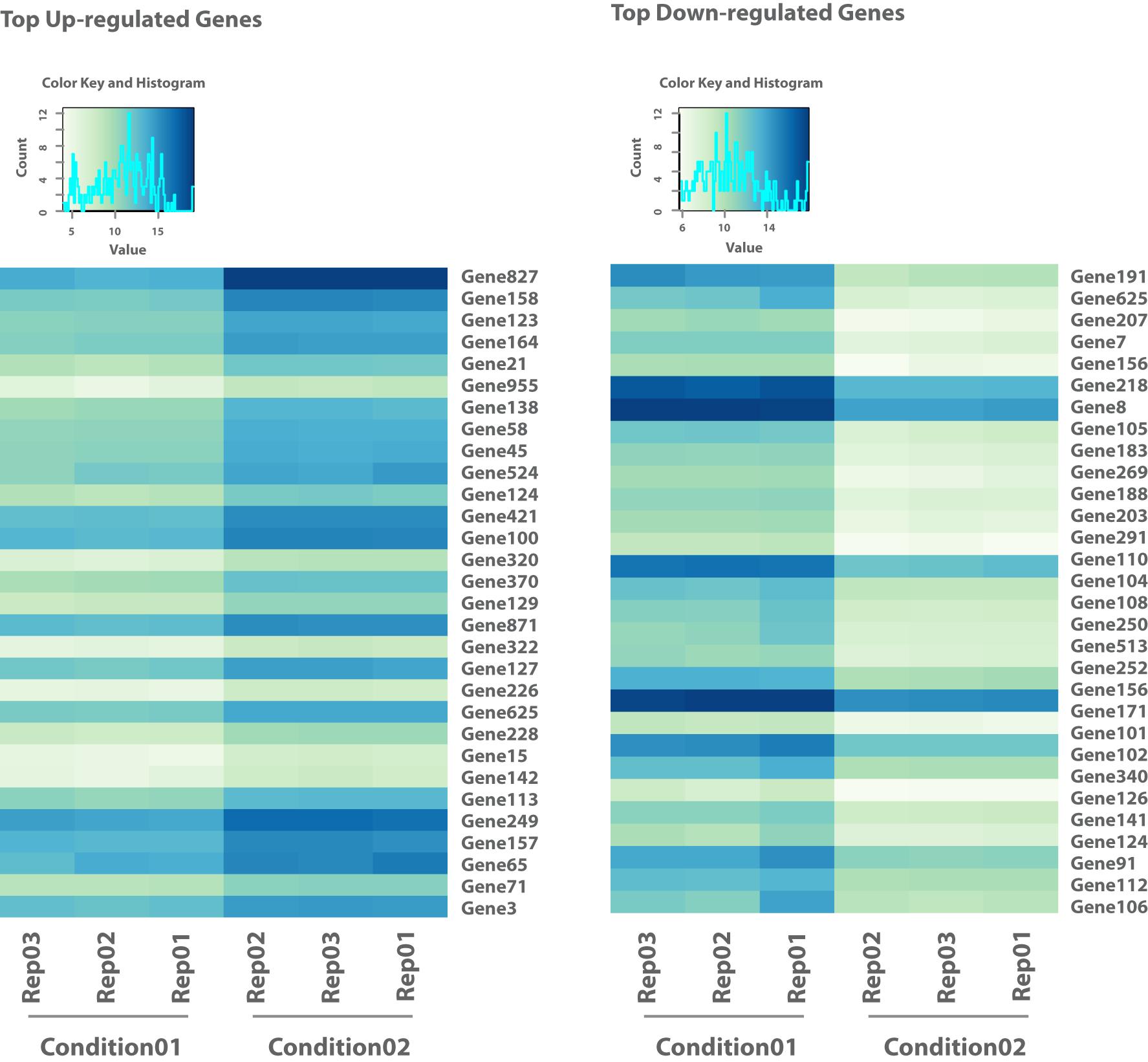
Obtenez les nombres de lectures mis en correspondance avec les caractéristiques annotées telles que les gènes ou les transcrits pour une analyse quantitative approfondie. - Visualisation de l'analyse statistique (par exemple au format .png, voir figure 1-5)
Accédez à des représentations visuellement informatives des analyses statistiques, y compris des diagrammes en forme de volcan, des cartes thermiques, des voies enrichies et des cartes d'exon.
Exclusif aux transcriptomes eucaryotes :
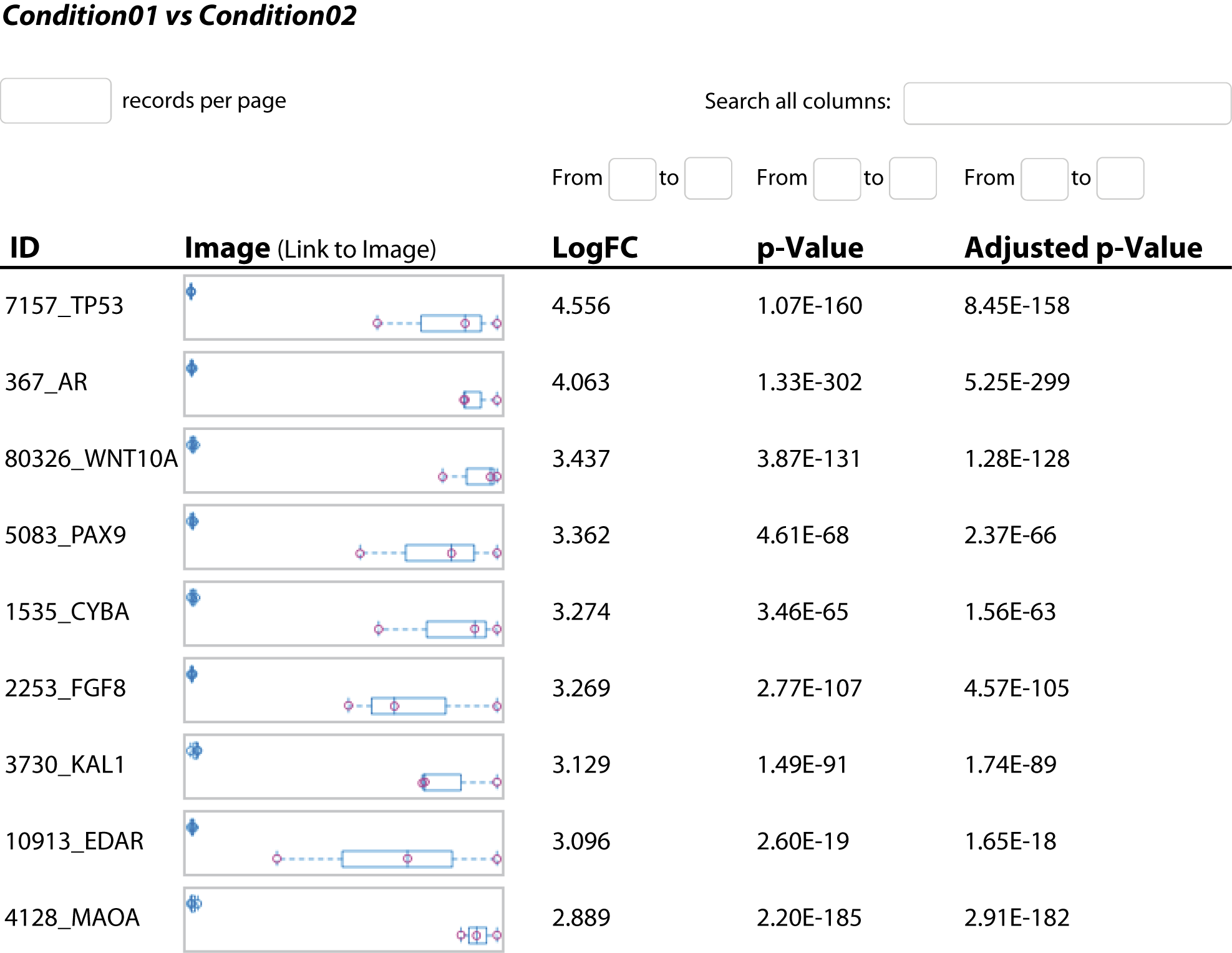
- Analyse de l'expression génique différentielle (en format .tsv et .html interactif, voir figure 3)
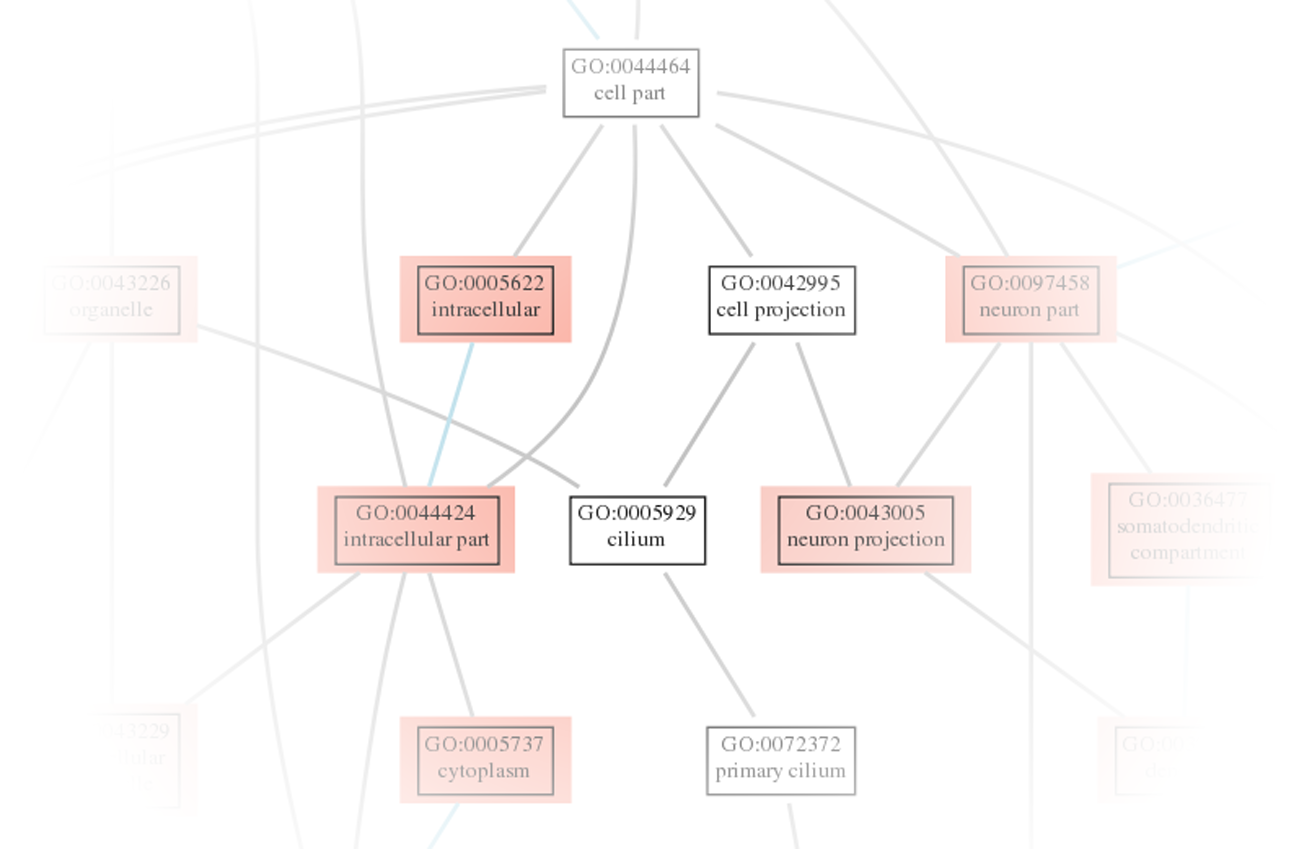
Explorez l'expression différentielle des gènes grâce à des résultats complets sous forme de tableaux et de fichiers HTML. - Analyse des jeux de gènes (en format .tsv et .html, voir figure 4)
Découvrez les voies enrichies grâce à l'analyse des jeux de gènes, disponible en formats tabulaire et HTML. - Analyse de l'utilisation différentielle des exons (en format .tsv et .html, voir figure 5)
Découvrez l'épissage alternatif grâce à l'analyse de l'utilisation différentielle des exons, dont les résultats sont fournis sous forme de tableaux et de fichiers HTML.
Ces produits livrables fournissent des données exploitables à partir de votre analyse de séquençage de l'ARNm, permettant une prise de décision éclairée.
Analyse bioinformatique complémentaire (moyennant des frais supplémentaires)
- Détermination des variants SNV et des petits InDels (<50bp) dans le transcriptome (au format .vcf)
Identifier les variants de nucléotides simples (SNV) et les petites insertions/délétions (InDels) dans le transcriptome grâce aux résultats de l'appel de variants fournis au format .vcf.
Avec la configuration expérimentale appropriée et la disponibilité de données publiques pertinentes, ces services supplémentaires offrent une compréhension plus approfondie de vos échantillons.
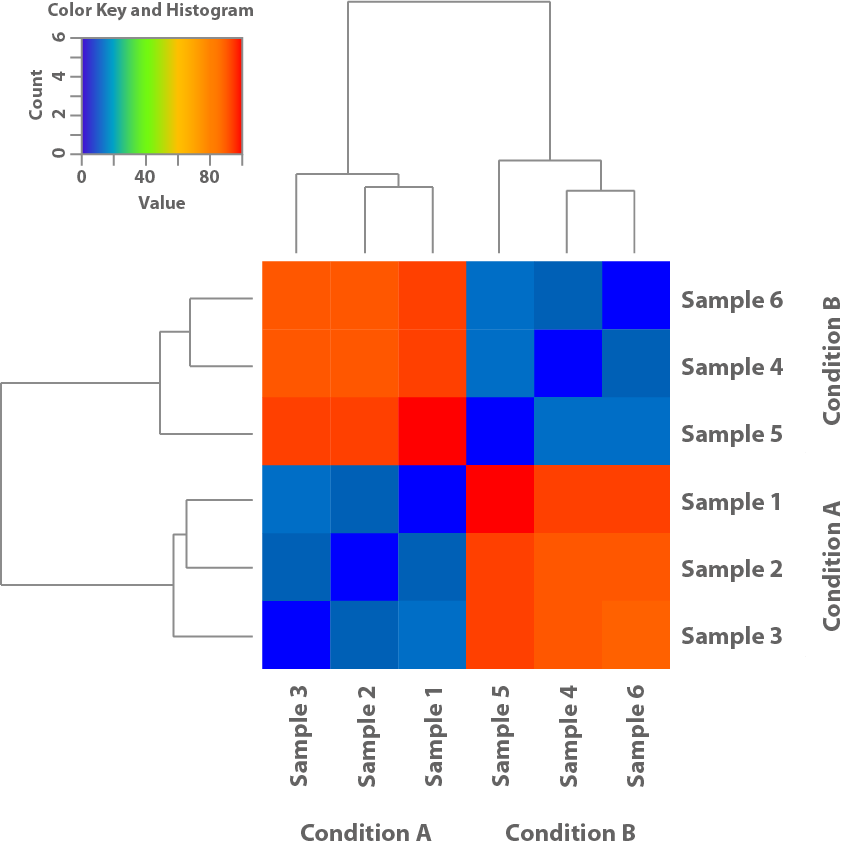
Figure 1: Cette heatmap est basée sur les modèles d'expression des échantillons et montre leur similarité les uns par rapport aux autres. Elle permet ainsi de déterminer si les conditions utilisées dans l'expérience conduisent à des schémas d'expression différents.
Délai d'Exécution
- Livraison des données dans les 25 jours ouvrables suivant la réception de l'échantillon (y compris la préparation de la bibliothèque et le séquençage)
- 5 jours ouvrables supplémentaires pour l'analyse des données (bioinformatique)
- Service express possible sur demande